The recent success in getting at least some data from Phil Jones – which he had obstructed since my original request in 2003 – has caused me to refresh my attempts to get Lonnie Thompson to archive his data so that the scandalous inconsistencies between different versions can finally be appraised. Last year, he published an article drawing on seven tropical ice cores in PNAS, which has a data policy that provides inter alia:
Unique Materials: Authors must make Unique Materials (e.g., cloned DNAs; antibodies; bacterial, animal, or plant cells; viruses; and computer programs) promptly available on request by qualified researchers for their own use.
and:
Databases: Before publication, authors must deposit large data sets (including microarray data, protein or nucleic acid sequences, and atomic coordinates for macromolecular structures) in an approved database and provide an accession number for inclusion in the published paper. When no public repository exists, authors must provide the data as Supporting Information online or, in special circumstances when this is not possible, on the author’s institutional web site, provided that a copy of the data is provided to PNAS.
These policies seem a little better on paper than some other journal policies, In addition, their webpage invites people experiencing problems to write to them. So I did so. Here’s my letter:
Dear Sirs,
Last year, I was invited to make a presentation to the National Academy of Sciences Panel on Surface Temperature Reconstructions on millennial temperature reconstructions and have published several peer-reviewed articles in the field, which were cited by the above panel in their report last year.
I am writing in connection pursuant to your policies for availability of unique materials and databases http://www.pnas.org/misc/iforc.shtml#submission in connection with Thompson et al 2006, Abrupt tropical climate change: Past and present, PNAS 103, 10536-10543
Your policy statement says that:
Unique Materials: Authors must make Unique Materials (e.g., cloned DNAs; antibodies; bacterial, animal, or plant cells; viruses; and computer programs) promptly available on request by qualified researchers for their own use.
and:
Databases: Before publication, authors must deposit large data sets (including microarray data, protein or nucleic acid sequences, and atomic coordinates for macromolecular structures) in an approved database and provide an accession number for inclusion in the published paper. When no public repository exists, authors must provide the data as Supporting Information online or, in special circumstances when this is not possible, on the author’s institutional web site, provided that a copy of the data is provided to PNAS.
Thompson et al 2006 describe results from ice cores drilled at Dunde, Guliya, Dasuopu, Puruogangri, Quelccaya, Huascaran and Sajama. For each core, several thousand samples were taken and analyses on a sample-by-sample basis made for isotopes, chemistry and other indicators. The information for each core constitutes a large data set within the meaning of your policies. There is an excellent public repository for ice core data at the World Data Center for Paleoclimatology, which satisfies your definition of a public repository. Under your policies, Thompson et al had an obligation to archive this data as a condition of publication, but this appears to have been overlooked. Although Thompson et al provided a highly abbreviated summary of isotope information as Supplementary Information, the Supplementary Information is incomplete and not compliant with journal policies.
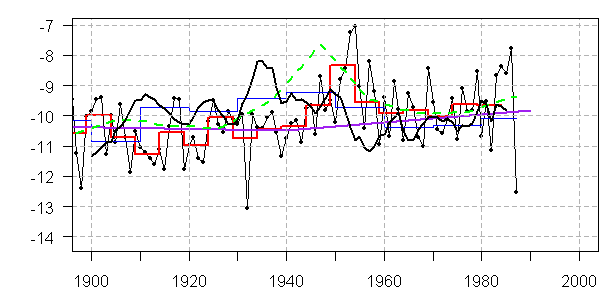
There is a pressing need to ensure compliance with journal data policies, because numerous inconsistent summaries are in gray and peer-reviewed circulation. For example, the figure below illustrates substantial differences between Dunde δO18 data summaries. These discrepancies can only be reconciled through examination of the underlying large data sets, which should have been archived prior to publication had journal policies been followed.
Dunde Versions. Heavy black — Yao et al 2006 (3 year rolling average); thin black – MBH98 (annual); red – PNAS 2006 (5-year averages); blue – Clim Chg 2003 (10-year averages); purple – Yang et al 2002 (values in 50 -year intervals); green – Crowley and Lowery 2000 (original in standardized format, re-fitted here for display by regression fit to MBH98).
I request that you ensure that Thompson et al comply with your data policy by forthwith archiving the large datasets used in the PNAS article for each individual ice core (Dunde, Dasuopu, Guliya, Puruoganri, Quelccaya, Sajama, Huascaran) and for the entire suite of isotopes and chemistry. In addition, because the discrepancies may result from changing algorithms for dating the ice cores, I further request that the dating procedure for each core be made available under your Unique Materials policy.
Thank you for your attention.
Stephen McIntyre
We’ll see what happens. BTW the National Academy panel on Data Integrity, promised in the wake of the Surface Temperature Panel, has been empanelled and held its first hearings this week. Gerry North was the first speaker. He’s sent me a copy of his presentation which I’ll post on some day.




18 Comments
This appears to be the NAS Panel referred to:
http://www8.nationalacademies.org/cp/projectview.aspx?key=48721
Am I the only one who could see #4 on that link being used against guys ike Steve?
I can see guys like Mann and Jones arguing that “denialists” like Steve and Willis are out here impugning the integrity of their research by “spreading misinformation on a non-peer reviewed website”.
#4 is interesting wording. I would argue that the only actions that we see here that “compromise the integrity of research data” are the actions taken by Jone, Mann, Thompson: losing the data, withholding adverse results,… What are the science community, journals etc doing about it?
The explicitly cited things for archiving are
and
All these things I duly archive, as do other biologists or population geneticists.
However, do I see any mentions of specific requests for archived tree rings, coordinates of meteostations, raw temp data including their processing or suchlikes? No…
Its funny how people can get themselves really worked up into a frenzy over some lost emails about why a couple of political appointees lost their jobs. But when it comes to years of data upon which global energy policy for the next 5 decades may be radically changed, it seems that “I lost it” is a perfectly acceptable answer. Strange times we live in…
EW, well, the “computer programs” part might be relevant. Was a program used to manipulate the data, to produce all those variations? If so, it seems their policy requires disclosure of the source code, and I would argue that if they don’t include the data it requires, the program is incomplete. Even if they argue that data is not part of the program (a dubious position), at least seeing what adjustments they made and how they made them would be valuable.
#4
If the wording were fashioned to carve out the hockey team/carbon trading interests, it would not include computer programs. Given the selected examples, I’d say the areas of medical and drug research are at the forefront of thinking.
In our context here. But given the other examples the threats to integrity that are most contemplated are shenanigans in the medical/drug contexts, say, the a suggestion to tweak or lose data, thereby its integrity. I didn’t take “integrity” to signal a concern about disclosure vs. non-disclosure.
#4 The PNAS covers a wide range of sciences. The PNAS data policy for databases and unique materials covers a wide range of databases and unique materials. The few items mentioned in the policy are clearly just examples, not an exhaustive list.
Steve’s letter to PNAS makes a compelling case that the large datasets and dating procedure used by Thompson et al 2006 PNAS article are subject to the PNAS data policy.
EW,
These journals could learn a lot from NCBI.
Sure. NCBI is great.
I think that the computer thing is included as an example again because of biologists – it’s all these programs for DNA/protein sequence analysis and comparisons and it’s well known that using the same method implemented in different programs might lead to slightly different phylogenies.Therefore archived datasets with parameters AND cited software are required.
That’s all very helpful. If they understand this in connection with biology, that makes an excellent precedent for paleoclimate.
I think it is a matter of record that in the earlier days of molecular biology, people would publish data on cloning something, and then not provide the sequence of the clone. This was sometimes because they could get another publication out of the sequence, and sometimes because they could monopolise all the research if they didn’t tell anyone else the sequence. This lead to all sorts of problems, because scientists could act irresponsibly, and frequently did. Thus the journals explicit guidance. It took a lot longer to get deposition of data coordinates for crystal structures, but that is now sufficiently common that all the journals simply mandate deposition in a databank. Again, people used to argue that they had spent a long time getting crystal structures, and that they should have privileged access to the structure for a year (or so…) before giving it to the community; this obviously lead to various sorts of anguish.
I suspect that the problem here is that you have a small community which is relatively closed and inbred, versus the biology community targeted above, which is large and outbred. The paleos will dislike the light of inquiry into their poor communal standards, because it is an obvious festering sore. However, you shouldn’t imagine that the biology community is exemplary; there are many examples where if you give someone the opportunity to hide stuff, they do so.
I would be very interested to see Gerry North’s presentation, or a link to it, if it ever becomes available publically. Given the remit of his committee, I would be fascinated to see how he would attack the problem.
per
#10,
Different algorithm on the same DNA data leading to different phylogenies has the equivalent in paleoclimate proxying variables called SSI (spaghetti sagging indexes)
Re #12 and hiding stuff. There is an amusing anecdote about Watson of DNA fame sending a letter to a competing lab asking for a sample of some bacteria or virus. He eventually got back a refusal letter. No problem, he had an assistant culture the letter. The internet is quick and sure but has its downside too.
#12. North sent me a copy of his presentation. I can’t see any basis for it being private, but I’ll ask him.
The PPT leads with an anti-Barton editorial from the Houston Chronicle and then as separate slides on both realclimate and climateaudit. ONe of his last slides is my letter post-NAS Panel to North asking for help getting data. (Not a single piece of data has been provided.)
Some of the points that North highlighted give a hugely misleading impression of data issues. For example, the NAS Panel was confronted by Hans von Storch condemning the Phil Jones refusal to identify data: We have 25 years invested in this, why should we let you see the data when your only objective is to find something wrong with it? Instead of this, North posted up a trivializing email exchange as follows:
As to the problems with tree ring data, he used the following quote:
Well, that’s true, but it doesn’t deal with the problem that important studies use unarchived data. It’s annoying and unfortunate that he mis-stated the issue.
His email to me said:
I find some of the emphasis on new policy to be frustrating. The US Global Change Research Program (which includes climate) already requires agencies to do ensure that data is archived – so there’s already a policy framework for climate without having to get involved in issues like biology, computers, all the problems that result from trying to develop a Napoleonic Code for data. I’d be content with a common law approach – case-by-case. If NSF simply did its job and ceased being co-opted by non-compliant scientists, most of the problems in paleoclimate would disappear instantly.
North said in his covering email that he’d said that the paleoclimate community was “shocked” to find themselves thrust into the limelight and “totally unprepared” for it. It’s funny – they didn’t seem to be shocked when they got awards from Scientific American or their results applied in Al Gore’s movie. What were they “unprepared for” – critical analysis of their results?
Re: #15
God help us if any of these scientists, like North, ever become real and elected politicians. They take obfuscation to a whole other level.
you may overestimate the science (and paleo) community. As you found with Jones data, scientists (who are not computer scientists) may simply fail to appreciate the problems with database integrity, and over-writing old data with new data- until they are ten years in and notice that their database is actually a pig’s ear of over-writes and non-overwrites. Once in that position, it can be very difficult to (a) own up that there is a problem (b) do something about it; grant authorities are unlikely to give you hundreds of thousands of $ because you have committed an elementary error.
I suspect Crowley’s problem of not having the original data, or not even knowing if you have the original data, is quite common, and not just in paleo.
All that you can do is shine light into dark and dusty corners, and reveal the horrors that lie therein. That is how GLP came about for pharmaceutical research, and is the basis for UK research council guidance now.
per
#4 —
If I understand correctly, your comment pertains to:
http://ancienthistory.about.com/od/abbreviations/f/ievseg.htm reads in part:
“i.e.
“I.e.” stands simply for “that is,” which written out fully in Latin is ‘id est’. “I.e.” is used in place of “in other words,” or “it/that is.” It specifies or makes more clear.
e.g.
“E.g.” means “for example” and comes from the Latin expression exempli gratia, “for the sake of an example,” with the noun exemplum in the genitive to go with gratia in the ablative. “E.g.” is used in expressions similar to “including,” when you are not intending to list everything that is being discussed.”
So, e.g. cannot be read to imply any limit to the list and therefore the policy statement can be read correctly without the paranthetical as:
“Unique Materials: Authors must make Unique Materials promptly available on request by qualified researchers for their own use.”
Any limitation to that blanket statement would have to be found in some other sentence.
5 Trackbacks
[…] data following his publication in Proceedings of the National Academy of Sciences see CA here. Despite clear policies that on their face require the archiving of large data sets, PNAS refused to […]
[…] this year on letters to NAS asking them to require Thompson to comply with their data policies. My initial inquiry got nowhere as noted […]
[…] sent me his presentation and gave me permission to post it up. My comments here are similar to my earlier comment. The PPT leads with an anti-Barton editorial from the Houston Chronicle and then as separate slides […]
[…] Himalayan ice core information for Dasupo, Dunde, and Gulaya, is detailed (inter alia) here, here, here, here, here, here, and […]
[…] I’ve been trying since 2003 to get detailed sample information from Lonnie Thompson on his tropical ice cores, some drilled 20 years ago. I reported on my most recent effort on Apr 19, 2007 under PNAS policies here. […]